
RESEARCH
METABOLIC NETWORKS
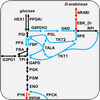
The metabolic network for an organism consists of all known metabolites and the enzymes that catalyze transformations between metabolites. By specifying a particular set of input conditions, such as glucose and oxygen uptake, we use mathematical models to determine the accumulation of biomass and, hence, the growth of a cell. We apply these models to study the metabolism of pathogens under different conditions and exploit these networks to determine drug-dose responses.
Publications
Pannala, V. R., M. R. Balik-Meisner, D. Mav, D. P. Phadke, E. H. Scholl, R. R. Shah, W. Casey, S. S. Auerbach, and A. Wallqvist. Fenofibrate differentially activates PPARα-mediated lipid metabolism in rat kidney and liver. Scientific Reports. 2025 October 22; 15:36842. [PDF, Pubmed]
Hari, A., M. R. Balik-Meisner, D. Mav, D. P. Phadke, E. H. Scholl, R. R. Shah, W. Casey, S. S. Auerbach, A. Wallqvist, and V. R. Pannala. Genome-scale metabolic modeling predicts per- and polyfluoroalkyl substance-mediated early perturbations in liver metabolism. Toxics. 2025 August 17; 13:684. [PDF, Pubmed]
Pannala, V. R., A. Hari, M. D. M. AbdulHameed, M. R. Balik-Meisner, D. Mav, D. P. Phadke, E. H. Scholl, R. R. Shah, S. S. Auerbach, and A. Wallqvist. Quantifying liver-toxic responses from dose-dependent chemical exposures using a rat genome-scale metabolic model. Toxicological Sciences. 2025 April; 204(2):154-168. [PDF, Pubmed]
Dougherty, B. V., C. J. Moore, K. D. Rawls, M. L. Jenior, B. Chun, S. Nagdas, J. J. Saucerman, G. L. Kolling, A. Wallqvist, and J. A. Papin. Identifying metabolic adaptations characteristic of cardiotoxicity using paired transcriptomics and metabolomics data integrated with a computational model of heart metabolism. PLoS Computational Biology. 2024 February 29; 20(2):e1011919. [PDF, Pubmed]
Tewari, S. G., R. Elahi, B. Kwan, K. Rajaram, S. Bhatnagar, J. Reifman, S. T. Prigge, A. B. Vaidya, and A. Wallqvist. Metabolic responses in blood-stage malaria parasites associated with increased and decreased sensitivity to PfATP4 inhibitors. Malaria Journal. 2023 February 14; 22:56. [PDF, Pubmed]
Rajaram, K., S. G. Tewari, A. Wallqvist, and S. T. Prigge. Metabolic changes accompanying the loss of fumarate hydratase and malate-quinone oxidoreductase in the asexual blood stage of Plasmodium falciparum. Journal of Biological Chemistry. 2022 May; 298(5):101897. [PDF, PubMed]
Tewari, S. G., B. Kwan, R. Elahi, K. Rajaram, J. Reifman, S. T. Prigge, A. B. Vaidya, and A. Wallqvist. Metabolic adjustments of blood-stage Plasmodium falciparum in response to sublethal pyrazoleamide exposure. Scientific Reports. 2022 January 21; 12(1):1167. [PDF, Pubmed]
Tewari, S. G., K. Rajaram, R. P. Swift, B. Kwan, J. Reifman, S. T. Prigge, and A. Wallqvist. Inter-study and time-dependent variability of metabolite abundance in cultured red blood cells. Malaria Journal. 2021 July 2; 20(1):299. [PDF, PubMed]
Tewari, S., K. Rajaram, R. Swift, J. Reifman, S. T. Prigge, and A. Wallqvist. Metabolic survival adaptations of Plasmodium falciparum exposed to sublethal doses of fosmidomycin. Antimicrobial Agents and Chemotherapy. 2021 March 18; 65(4):e02392-20. [PDF, PubMed]
Dougherty, B. V., K. D. Rawls, G. L. Kolling, K. C. Vinnakota, A. Wallqvist, and J. A. Papin. Identifying functional metabolic shifts in heart failure with the integration of omics data and a heart-specific, genome-scale model. Cell Reports. 2021 March 9; 34(10):108836. [PDF, PubMed]
Pannala, V. R., S. K. Estes, M. Rahim, I. Trenary, T. P. O’Brien, C. Shiota, R. L. Printz, J. Reifman, M. Shiota, J. D. Young, and A. Wallqvist. Toxicant-induced metabolic alterations in lipid and amino acid pathways are predictive of acute liver toxicity in rats. International Journal of Molecular Sciences. 2020 November 4; 21:8250. [PDF, PubMed]
Schyman, P., R. L. Printz, M. D. M. AbdulHameed, S. K. Estes, C. Shiota, M. Shiota, and A. Wallqvist. A toxicogenomic approach to assess kidney injury induced by mercuric chloride in rats. Toxicology. 2020 June 26; 442:152530. [PDF, PubMed]
Schyman, P., R. L. Printz, S. K. Estes, T. P. O'Brien, M. Shiota, and A. Wallqvist. Concordance between thioacetamide-induced liver injury in rat and human in vitro gene expression data. International Journal of Molecular Sciences. 2020 June 4; 21(11):4017. [PDF, PubMed]
Pannala, V. R., S. K. Estes, M. Rahim, I. Trenary, T. P. O'Brien, C. Shiota, R. L. Printz, J. Reifman, T. Oyama, M. Shiota, J. D. Young, and A. Wallqvist. Mechanism-based identification of plasma metabolites associated with liver toxicity. Toxicology. 2020 May 30; 441:152493. [PDF, PubMed]
Tewari, S. G., R. P. Swift, J. Reifman, S. T. Prigge, and A. Wallqvist. Metabolic alterations in the erythrocyte during blood-stage development of the malaria parasite. Malaria Journal. 2020 February 27; 19(1):94. [PDF, PubMed]
Swift, R. P., K. Rajaram, H. B. Liu, A. Dziedzic, A. E. Jedlicka, A. D. Roberts, K. A. Matthews, H. Jhun, N. N. Bumpus, S. G. Tewari, A. Wallqvist, and S. T. Prigge. A mevalonate bypass system facilitates elucidation of plastid biology in malaria parasites. PLOS Pathogens. 2020 February 14; 16(2):e1008316. [PDF, PubMed]
Pannala, V. R., K. C. Vinnakota, S. K. Estes, I. Trenary, T. P. O'Brien, R. L. Printz, J. A. Papin, J. Reifman, T. Oyama, M. Shiota, J. D. Young, and A. Wallqvist. Genome-scale model-based identification of metabolite indicators for early detection of kidney toxicity. Toxicological Sciences. 2020 February 1; 173(2):293-312. [PDF, PubMed]
Rawls, K. D., E. M. Blais, B. V. Dougherty, K. C. Vinnakota, V. R. Pannala, A. Wallqvist, G. L. Kolling, and J. A. Papin. Genome-scale characterization of toxicity-induced metabolic alterations in primary hepatocytes. Toxicological Sciences. 2019 December 1; 172(2):279-291. [PDF, PubMed]
Stewart, P. S., B. White, L. Boegli, T. Hamerly, K. S. Williamson, M. J. Franklin, B. Bothner, G. A. James, S. Fisher, F. G. Vital-Lopez, and A. Wallqvist. Conceptual model of biofilm antibiotic tolerance that integrates phenomena of diffusion, metabolism, gene expression, and physiology. Journal of Bacteriology. 2019 November 15; 201(22):e00307-19. [PDF, PubMed]
Pannala, V. R., K. C. Vinnakota, K. D. Rawls, S. K. Estes, T. P. O'Brien, R. L. Printz, J. A. Papin, J. Reifman, M. Shiota, J. D. Young, and A. Wallqvist. Mechanistic identification of biofluid metabolite changes as markers of acetaminophen-induced liver toxicity in rats. Toxicology and Applied Pharmacology. 2019 June 1; 372:19-32. [PDF, PubMed]
Tewari, S. G., K. Rajaram, P. Schyman, R. Swift, J. Reifman, S. T. Prigge, and A. Wallqvist. Short-term metabolic adjustments in Plasmodium falciparum counter hypoxanthine deprivation at the expense of long-term viability. Malaria Journal. 2019 March 19; 18:86. [PDF, PubMed]
Vinnakota, K. C., V. R. Pannala, M. L. Wall, M. Rahim, S. K. Estes, I. Trenary, T. P. O'Brien, R. L. Printz, J. Reifman, M. Shiota, J. D. Young, and A. Wallqvist. Network modeling of liver metabolism to predict plasma metabolite changes during short-term fasting in the laboratory rat. Frontiers in Physiology. 2019 March 1; 10:161. [PDF, PubMed]
Rawls, K. D., B. V. Dougherty, E. M. Blais, E. Stancliffe, G. L. Kolling, K. Vinnakota, V. R. Pannala, A. Wallqvist, and J. A. Papin. A simplified metabolic network reconstruction to promote understanding and development of flux balance analysis tools. Computers in Biology and Medicine. 2019 February; 105:64-71. [PDF, PubMed]
Pannala, V. R., M. L. Wall, S. K. Estes, I. Trenary, T. P. O’Brien, R. L. Printz, K. C. Vinnakota, J. Reifman, M. Shiota, J. D. Young, and A. Wallqvist. Metabolic network-based predictions of toxicant-induced metabolite changes in the laboratory rat. Scientific Reports. 2018 August 3; 8:11678. [PDF, PubMed]
Tewari, S. G., S. T. Prigge, J. Reifman, and A. Wallqvist. Using a genome-scale metabolic network model to elucidate the mechanism of chloroquine action in Plasmodium falciparum. International Journal for Parasitology: Drugs and Drug Resistance. 2017 March 22; 7(2):138-146. [PDF, PubMed]
Blais, E. M., K. D. Rawls, B. V. Dougherty, Z. I. Li, G. L. Kolling, P. Ye, A. Wallqvist, and J. A. Papin. Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nature Communications. 2017 February 8; 8:14250. [PDF, PubMed]
Wallqvist, A., X. Fang, S. G. Tewari, P. Ye, and J. Reifman. Metabolic host responses to malarial infection during the intraerythrocytic developmental cycle. BMC Systems Biology. 2016 August 8; 10:58. [PDF, PubMed]
Vital-Lopez, F. G., J. Reifman, and A. Wallqvist. Biofilm formation mechanisms of Pseudomonas aeruginosa predicted via genome-scale kinetic models of bacterial metabolism. PLOS Computational Biology. 2015 October 2; 11(10):e1004452. [PDF, PubMed]
Song, H. S., J. Reifman, and A. Wallqvist. Prediction of metabolic flux distribution from gene expression data based on the flux minimization principle. PLOS ONE. 2014 November 14; 9(11):e112524. [PDF, PubMed]
Fang, X., J. Reifman, and A. Wallqvist. Modeling metabolism and stage-specific growth of Plasmodium falciparum HB3 during the intraerythrocytic developmental cycle. Molecular BioSystems. 2014 October 1; 10:2526-2537. [PDF, PubMed]
Vital-Lopez, F. G., A. Wallqvist, and J. Reifman. Bridging the gap between gene expression and metabolic phenotype via kinetic models. BMC Systems Biology. 2013 July 22; 7:63. [PDF, PubMed]
Chaudhury, S., M. D. AbdulHameed, N. Singh, G. Tawa, P. M. D'haeseleer, A. T. Zemla, A. Navid, C. E. Zhou, M. C. Franklin, J. Cheung, M. J. Rudolph, J. Love, J. F. Graf, D. A. Rozack, J. L. Dankmeyer, K. Amemiya, S. Daefler, and A. Wallqvist. Rapid countermeasure discovery against Francisella tularensis based on a metabolic network reconstruction. PLOS ONE. 2013 May 21; 8(5):e63369. [PDF, PubMed]
Fang, X., A. Wallqvist, and J. Reifman. Modeling phenotypic metabolic adaptations of Mycobacterium tuberculosis H37Rv under hypoxia. PLOS Computational Biology. 2012 September 13; 8(9):e1002688. [PDF, PubMed]
Fang, X., A. Wallqvist, and J. Reifman. Modeling synergistic drug inhibition of Mycobacterium tuberculosis growth in murine macrophages. Molecular BioSystems. 2011 September 1; 7(9):2622-2636. [PDF, PubMed]
Fang, X., A. Wallqvist, and J. Reifman. Development and analysis of an in vivo-compatible metabolic network of Mycobacterium tuberculosis. BMC Systems Biology. 2010 November 26; 4:160. [PDF, PubMed]
Fang, X., A. Wallqvist, and J. Reifman. A systems biology framework for modeling metabolic enzyme inhibition of Mycobacterium tuberculosis. BMC Systems Biology. 2009 September 15; 3:92. [PDF, PubMed]