
RESEARCH
DRUG DESIGN & MOLECULAR INTERACTIONS
We combine the power of DoD high-performance computing with molecular modeling techniques to develop tools to evaluate how small molecules interact with biological macromolecules. We aim to develop and repurpose drugs, ascertain adverse drug effects, predict protein structures and consequences of amino acid mutations, and model biological and chemical interactions at membranes. We use these methods to understand chemical and biological processes at the atomistic and molecular levels and to exploit this knowledge in drug design.
Research Projects
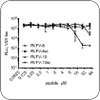 |
Drug Development
These projects combine computational and experimental efforts to repurpose drugs; develop drugs and vaccines against toxins, bacteria, and viruses; and counter drug resistance. |
 |
Membrane Systems & Antimicrobial Peptides
This research uses atomistic and coarse-grained models of lipid bilayers and antimicrobial peptides to simulate biological processes at cellular membranes. |
 |
Protein Engineering
This research addresses how the structural and dynamical properties of proteins and peptides affect their behavior and biological functions. |
 |
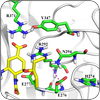
Protein Structure Predictions
We use custom methods that predict the structures of novel amino acid sequences to screen drugs, assess vaccines, understand reactions, and model molecular interactions. |
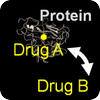 |
Repurposing & Adverse Effects
We use patterns of drug-protein interactions to identify drug repurposing candidates and adverse effects of drug-drug interactions. |
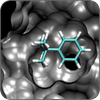 |
Small-Molecule Protein Interactions
We model how drugs and substrates bind to their protein targets, allowing us to select and modify compounds with improved efficacies. |