
RESEARCH
BIOINFORMATICS & GENOMICS
We develop and deploy a range of bioinformatics software pipelines to DoD high-performance computing assets for annotating whole genome function, developing unique DNA fingerprints for pathogens, and comparing genomes. Research topics include, creating diagnostic platforms for detecting and strain typing pathogens, understanding bacterial secretion systems and host-pathogen interactions, investigating the human microbiome, predicting orthology, revealing thermodynamic forces underlying viral evolution, and analyzing genotype-phenotype associations.
Research Projects
 |
Comparative Genomics
We develop high-throughput computational approaches to enable comparative genomics studies based on orthology. |
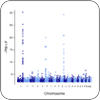 |
Genotype-Phenotype Analysis
We develop algorithms for inferring disease risks from genomic data and apply them to better undertand human susceptibility to traumatic events and sleep deprivation. |
 |
Host-Pathogen Interactions
We identify virulence factors associated with bacterial secretion systems via bioinformatics and in vitro and in vivo experimentation. |
 |
Pathogen Diagnostics
We design customized and platform-specific diagnostic probe sets to identify known and new pathogen species and strains. |
 |
Respiratory Microbiome
We investigate how alterations in the respiratory microbiome of Service members affect their health and factors associated with chronic and acute respiratory disease. |
 |
Sequencing and Annotation
We develop high-throughput computational solutions for annotating, analyzing, managing, and storing genome-scale sequencing data. |
 |
Viral Evolution
Using sequencing data and mathematical models, we study the capacity of viral pathogens to create escape mutants that evade the host immune system. |